# SPDX-License-Identifier: MIT
# Copyright (c) 2018 University of Zurich
Visualizer¶
Model: The model represents the data, and does nothing else. The model does NOT depend on the controller or the view.
EventsModel
StateVariablesModel
…
View: The view displays the model data, and sends user actions (e.g. button clicks) to the controller. The view can be independent of both the model and the controller or actually be the controller, and therefore depend on the model.
HistogramViewer
RasterplotViewer
LineplotViewer
…
Controller: The controller provides model data to the view, and interprets user actions such as button clicks. The controller depends on the view and the model. In some cases, the controller and the view are the same object.
HistogramController (called Histogram)
RasterplotController (called Rasterplot)
LineplotController (called Lineplot)
…
- PlotSettings()
font sizes
…
Backends Currently, the visualizer supports matplotlib and pyqtgraph as backends. To switch between the two backends you simply have to change one input flag of the controller. This is shown below in the examples which are given for pyqtgraph and for matplotlib, however the figure are only shown for matplotlib.
How to use it - in short¶
1) get data
spikemonN1, spikemonN2, statemonN1, statemonN2 = run_your_own_brian_network()
2) define PLOTSETTING
from teili.tools.visualizer.DataViewers import PlotSettings
MyPlotSettings = PlotSettings(fontsize_title=20, fontsize_legend=14,
Fontsize_axis_labels=14, marker_size = 30,colors = [‘r’, ‘b’, ‘g’, ‘c’,
‘k’, ‘m’, ‘y’])
3) call CONTROLLER of desired type of plot, e.g. Histogram, Rasterplot, Lineplot, …
from teili.tools.visualizer.DataControllers import Histogram
DataModel_to_attr = [(spikemonN1, ‘i’), (spikemonN2, ‘i’)]
subgroup_labels = [‘N1’, ‘N2’]
HC = Histogram(MyPlotSettings=MyPlotSettings, DataModel_to_attr=DataModel_to_attr, subgroup_labels=subgroup_labels, backend=‘matplotlib’)
If you want to change anything else on the main figure/window or one of the subplots you can directly access the mainfigure (matplotlib: figure, pyqtgraph: qt-window) as my_controller.mainfig or the corresponding subplot under my_controller.subfig
How to use it - the slightly longer version¶
Import the required modules and functions …
%pylab inline
import numpy as np
import os
from brian2 import us, ms, second, prefs, defaultclock, start_scope, SpikeGeneratorGroup, SpikeMonitor, StateMonitor
import matplotlib.pylab as plt
import pyqtgraph as pg
from PyQt5 import QtGui
from teili.core.groups import Neurons, Connections
from teili import TeiliNetwork
from teili.models.neuron_models import DPI
from teili.models.synapse_models import DPISyn
from teili.models.parameters.dpi_neuron_param import parameters as neuron_model_param
from teili.models.parameters.dpi_synapse_param import parameters as synapse_model_param
QtApp = QtGui.QApplication([])
def run_brian_network():
prefs.codegen.target = "numpy"
defaultclock.dt = 10 * us
start_scope()
N_input, N_N1, N_N2 = 1, 5, 3
duration_sim = 100
Net = TeiliNetwork()
# setup spike generator
spikegen_spike_times = np.sort(np.random.choice(size=500, a=np.arange(float(defaultclock.dt), float(duration_sim*ms)*0.9,
float(defaultclock.dt*5)), replace=False)) * second
spikegen_neuron_ids = np.zeros_like(spikegen_spike_times) / ms
gInpGroup = SpikeGeneratorGroup(N_input, indices=spikegen_neuron_ids,
times=spikegen_spike_times, name='gtestInp')
# setup neurons
testNeurons1 = Neurons(N_N1, equation_builder=DPI(num_inputs=2), name="testNeuron")
testNeurons1.set_params(neuron_model_param)
testNeurons2 = Neurons(N_N2, equation_builder=DPI(num_inputs=2), name="testNeuron2")
testNeurons2.set_params(neuron_model_param)
# setup connections
InpSyn = Connections(gInpGroup, testNeurons1, equation_builder=DPISyn(), name="testSyn", verbose=False)
InpSyn.connect(True)
InpSyn.weight = '200 + rand() * 100'
Syn = Connections(testNeurons1, testNeurons2, equation_builder=DPISyn(), name="testSyn2", verbose=False)
Syn.connect(True)
Syn.weight = '200 + rand() * 100'
# spike monitors input and network
spikemonInp = SpikeMonitor(gInpGroup, name='spikemonInp')
spikemonN1 = SpikeMonitor(testNeurons1, name='spikemon')
spikemonN2 = SpikeMonitor(testNeurons2, name='spikemonOut')
# state monitor neurons
# statemonN1 = StateMonitor(testNeurons1, variables=["Iin", "Imem"], record=[0, 3], name='statemonNeu')
statemonN1 = StateMonitor(testNeurons1, variables=["Iin", "Iahp"], record=True, name='statemonNeu')
# statemonN2 = StateMonitor(testNeurons2, variables=['Iahp'], record=0, name='statemonNeuOut')
statemonN2 = StateMonitor(testNeurons2, variables=['Imem'], record=True, name='statemonNeuOut')
Net.add(gInpGroup, testNeurons1, testNeurons2, InpSyn, Syn, spikemonN1, spikemonN2, statemonN1, statemonN2)
# run simulation
Net.run(duration_sim * ms)
print ('Simulation run for {} ms'.format(duration_sim))
return spikemonN1, spikemonN2, statemonN1, statemonN2
Get the data to plot¶
Option A: run brian network to get SpikeMonitors and StateMonitors
spikemonN1, spikemonN2, statemonN1, statemonN2 = run_brian_network()
Option B: create DataModel instance from arrays, lists or brian-SpikeMonitors/StateMonitors
Available DataModels:
EventsModel: stores neuron_ids and spike_times
Example shows how to create it from array/list …
from teili.tools.visualizer.DataModels import EventsModel
neuron_ids = [1, 1, 1, 2, 3, 1, 4, 5]
spike_times = [11, 14, 14, 16, 17, 25, 36, 40]
EM = EventsModel(neuron_ids=neuron_ids, spike_times=spike_times)
… or create from brian spike monitor
EM = EventsModel.from_brian_spike_monitor(spikemonN1)
Then the created EventsModel EM has the following attributes:
neuron_ids :
[3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4
1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 2 3 4 1 0 3 2 4 1 3 0 2 4 1 3
0 2 4 1 3 0 2 4 1 3 0 2 4 3 1 0 2 4 3 1 0 2 4 3 1 0 2 4 3 1 0 2 4 3 1 0 2
4 3 1 0 2 4 3 1 0 2 4 3 1 0 2 3 4 1 0 2 3 4 1 0 3 2 4 1 0 3 4 2 1 3 0 4 2
1 3 0 4 2 1 3 0 4 2 1 3 0 4 2 1 3 4 0 2 1 3 4 0 2 1 3 4 0 2 1 3 4 0 2 3 1
4 0 2 3 1 4 0 2 3 1 4 0 3 2 1 4 0 3 2 1]
spike_times :
[0.00387 0.004 0.00405 0.00411 0.00413 0.00628 0.00651 0.00659 0.00669
0.00673 0.0085 0.0088 0.00891 0.00903 0.00908 0.0107 0.0111 0.01124
0.01139 0.01145 0.01278 0.01326 0.01344 0.01363 0.0137 0.01491 0.01552
0.01574 0.01595 0.01603 0.01699 0.01764 0.01788 0.01812 0.01821 0.01907
0.01984 0.02012 0.02039 0.02049 0.02108 0.02183 0.02216 0.02247 0.02259
0.02304 0.02391 0.02426 0.0246 0.02473 0.02506 0.02604 0.02644 0.02684
0.027 0.02719 0.02822 0.02867 0.02914 0.02932 0.02935 0.03047 0.03094
0.03139 0.03144 0.03155 0.03261 0.03313 0.03351 0.03359 0.03375 0.0347
0.03525 0.03554 0.03576 0.03594 0.03682 0.03731 0.03748 0.03781 0.038
0.03885 0.03937 0.03945 0.03987 0.04005 0.04084 0.04141 0.04142 0.042
0.04222 0.04304 0.04359 0.04372 0.04438 0.04459 0.04526 0.04569 0.04591
0.04663 0.04688 0.04752 0.04786 0.04816 0.04878 0.04902 0.04953 0.0498
0.05021 0.0509 0.05117 0.05161 0.0518 0.05233 0.05309 0.05338 0.05373
0.05384 0.05449 0.05522 0.0555 0.05575 0.05579 0.05655 0.05743 0.05774
0.05788 0.05792 0.0587 0.05949 0.05978 0.05978 0.05989 0.06072 0.06163
0.0618 0.06194 0.06198 0.06289 0.06389 0.06391 0.06419 0.06423 0.06514
0.06605 0.06618 0.06643 0.06656 0.06748 0.06828 0.06855 0.06871 0.0689
0.06966 0.07033 0.07075 0.07086 0.07114 0.07195 0.0725 0.07303 0.07306
0.07343 0.07423 0.07471 0.0754 0.07545 0.0759 0.07656 0.07688 0.07763
0.07774 0.07815 0.07875 0.07899 0.07997 0.08017 0.08062 0.0811 0.08119
0.08212 0.08238 0.08286 0.08328 0.0833 0.08428 0.08459 0.08509 0.08537
0.0855 0.08647 0.08687 0.08744 0.08757 0.08783 0.08879 0.08929 0.08982
0.08987 0.0902 0.09128 0.0921 0.09261 0.09306 0.09351]
StateVariablesModel: stores any number of variables with their name and the list of timepoints when the variable was sampled
Example shows how to create it from array/list or from brian spike monitor
from teili.tools.visualizer.DataModels import StateVariablesModel
# create from array/list
state_variable_names = ['var_name']
num_neurons, num_timesteps = 6, 50
state_variables = [np.random.random((num_neurons, num_timesteps))]
state_variables_times = [np.linspace(0, 100, num_timesteps)]
SVM = StateVariablesModel(state_variable_names, state_variables, state_variables_times)
# create from brian state monitorS
skip_not_rec_neuron_ids=False
SVM = StateVariablesModel.from_brian_state_monitors([statemonN1, statemonN2], skip_not_rec_neuron_ids)
skip_not_rec_neuron_ids=True
SVM = StateVariablesModel.from_brian_state_monitors([statemonN1, statemonN2], skip_not_rec_neuron_ids)
Then the created StateVariablesModel SVM has the following attributes:
Iin :
[[0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
0.00000000e+00]
[0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
0.00000000e+00]
[0.00000000e+00 0.00000000e+00 0.00000000e+00 0.00000000e+00
0.00000000e+00]
...
[6.82521123e-09 7.02939025e-09 6.74769896e-09 7.76629202e-09
7.21872104e-09]
[6.81237889e-09 7.01617406e-09 6.73501234e-09 7.75169045e-09
7.20514890e-09]
[6.79957068e-09 7.00298271e-09 6.72234958e-09 7.73711634e-09
7.19160228e-09]]
t_Iin :
[0.000e+00 1.000e-05 2.000e-05 ... 9.997e-02 9.998e-02 9.999e-02]
Iahp :
[[5.00000000e-13 5.00000000e-13 5.00000000e-13 5.00000000e-13
5.00000000e-13]
[5.00000000e-13 5.00000000e-13 5.00000000e-13 5.00000000e-13
5.00000000e-13]
[5.00000000e-13 5.00000000e-13 5.00000000e-13 5.00000000e-13
5.00000000e-13]
...
[2.35349697e-11 2.45322975e-11 2.37997191e-11 2.54116963e-11
2.38412778e-11]
[2.35283328e-11 2.45253794e-11 2.37930076e-11 2.54045302e-11
2.38345545e-11]
[2.35216978e-11 2.45184633e-11 2.37862979e-11 2.53973662e-11
2.38278332e-11]]
t_Iahp :
[0.000e+00 1.000e-05 2.000e-05 ... 9.997e-02 9.998e-02 9.999e-02]
Imem :
[[0.00000000e+00 0.00000000e+00 0.00000000e+00]
[4.74578721e-33 4.74578721e-33 4.74578721e-33]
[9.49157441e-33 9.49157441e-33 9.49157441e-33]
...
[1.14559533e-10 2.80317027e-10 3.29995059e-10]
[1.15005619e-10 2.80084652e-10 3.29576244e-10]
[1.15447969e-10 2.79851599e-10 3.29157605e-10]]
t_Imem :
[0.000e+00 1.000e-05 2.000e-05 ... 9.997e-02 9.998e-02 9.999e-02]
Plot the collected data¶
Define PlotSettings¶
The PlotSettings are defined only once for all the plots that will be created. This should make it easier to get consistent color-codings, fontsizes and markersize across different plots.
The colors can be defined as RGBA to additionally define the transparency
from teili.tools.visualizer.DataViewers import PlotSettings
MyPlotSettings = PlotSettings(fontsize_title=20, fontsize_legend=14, fontsize_axis_labels=14,
marker_size = 30, # default 5
colors = ['r', 'b'], # default ['r', 'b', 'g', 'c', 'k', 'm', 'y']
)
Call the DataController of the desired type of plot¶
- So far in teili:
Histogram
Rasterplot
Lineplot
Histogram¶
Histogram - Inputs
* DataModel_to_attr --> e.g. [(spikemonN1, 'i'), (spikemonN2, 'i')] OR
[(EventsModel, 'i'), (EventsModel, 'i')]
* MyPlotSettings=PlotSettings()
* subgroup_labels=None --> e.g. ['Neurongroup N1', 'Neurongroup N2']
* bins=None --> e.g. range(0,9)
* orientation='vertical' --> 'horizontal' OR 'vertical'
* title='histogram
* xlabel='bins'
* ylabel='count',
* backend='matplotlib'
* show_immediately=False
Simple example to plot a histogram of two NeuronGroups to plot data from BrianSpikeMontiors/StateMonitors (or Datamodels) with matplotlib backend:
from teili.tools.visualizer.DataControllers import Histogram
# brian spike monitor
DataModel_to_attr = [(spikemonN1, 'i'), (spikemonN2, 'i')]
# or plot data from DataModels
# EM1 = EventsModel.from_brian_spike_monitor(spikemonN1)
# EM2 = EventsModel.from_brian_spike_monitor(spikemonN2)
# DataModel_to_attr = {EM1: 'neuron_ids', EM2:'neuron_ids'}
subgroup_labels = ['N1', 'N2']
HC = Histogram(DataModel_to_attr=DataModel_to_attr,
MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='matplotlib')
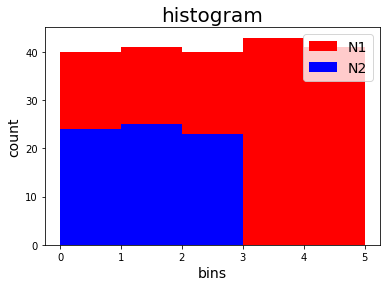
or PYQTGRAPH backend:
HC = Histogram(DataModel_to_attr=DataModel_to_attr,
MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='pyqtgraph',
QtApp=QtApp, show_immediately=True)
Rasterplot¶
Rasterplot - Inputs
* MyEventsModels --> list of EventsModel or BrianSpikeMonitors
* MyPlotSettings=PlotSettings()
* subgroup_labels=None --> ['N1', 'N2']
* time_range=None --> (0, 0.9)
* neuron_id_range=None, --> (0, 4)
* title='raster plot'
* xlabel='time
* ylabel='count',
* backend='matplotlib'
* add_histogram=False --> show histogram of spikes per neuron id next to rasterplot
* show_immediately=False
Simple example to plot a raster plot of two NeuronGroups to plot data from BrianSpikeMontiors/StateMonitors (or Datamodels) with matplotlib backend:
from teili.tools.visualizer.DataControllers import Rasterplot
# plot data from BrianSpikeMontiors
MyEventsModels = [spikemonN1, spikemonN2]
# or plot data from EventsModel
# EM1 = EventsModel.from_brian_spike_monitor(spikemonN1)
# EM2 = EventsModel.from_brian_spike_monitor(spikemonN2)
# MyEventsModels = [EM1, EM2]
subgroup_labels = ['N1', 'N2']
# MATPLOTLIB backend - WITHOUT HISTOGRAM
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels, backend='matplotlib')
# MATPLOTLIB backend - WITH HISTOGRAM
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels, add_histogram=True)
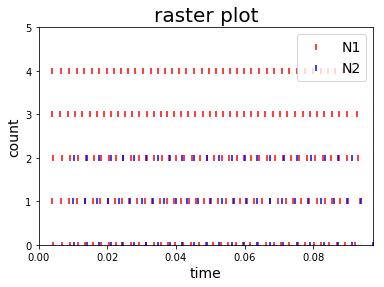
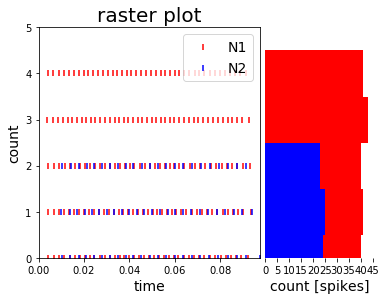
or PYQTGRAPH backend:
# PYQTGRAPH backend - WITHOUT HISTOGRAM
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels, backend='pyqtgraph', QtApp=QtApp)
# PYQTGRAPH backend - WITH HISTOGRAM
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels,
add_histogram=True, backend='pyqtgraph', QtApp=QtApp, show_immediately=True)
LinePlot¶
Lineplot - Inputs
* DataModel_to_x_and_y_attr --> e.g. [(statemonN1, ('Imem', 't_Imem')),
(statemonN2, ('Iahp', 't_Iahp'))]
OR
[(StateVariablesModel_N1, ('Imem', 't_Imem')),
(StateVariablesModel_N2, ('Iahp', 't_Iahp'))]
* MyPlotSettings=PlotSettings()
* subgroup_labels=None --> ['N1', 'N2']
* x_range=None, --> (0, 0.9)
* y_range=None, --> (0, 4)
* title='Lineplot'
* xlabel=None
* ylabel=None
* backend='matplotlib'
* show_immediately=False
Simple example to plot a line plot of two NeuronGroups to plot data from BrianSpikeMontiors/StateMonitors (or Datamodels) with matplotlib backend:
from teili.tools.visualizer.DataControllers import Lineplot
# plot data from BrianSpikeMontiors
DataModel_to_x_and_y_attr = [(statemonN1, ('t', 'Iin')), (statemonN2, ('t', 'Imem'))]
# or plot data from StateVariablesModel
SVM_N1 = StateVariablesModel.from_brian_state_monitors([statemonN1])
SVM_N2 = StateVariablesModel.from_brian_state_monitors([statemonN2])
DataModel_to_x_and_y_attr = [(SVM_N1, ('t_Iin', 'Iin')), (SVM_N2, ('t_Imem', 'Imem'))]
subgroup_labels = ['N1', 'N2']
LC = Lineplot(DataModel_to_x_and_y_attr=DataModel_to_x_and_y_attr,
MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='matplotlib')
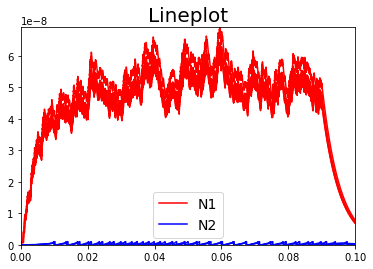
or PYQTGRAPH backend:
LC = Lineplot(DataModel_to_x_and_y_attr=DataModel_to_x_and_y_attr,
MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='pyqtgraph', QtApp=QtApp, show_immediately=True)
Additional functionalities¶
Combine different plots¶
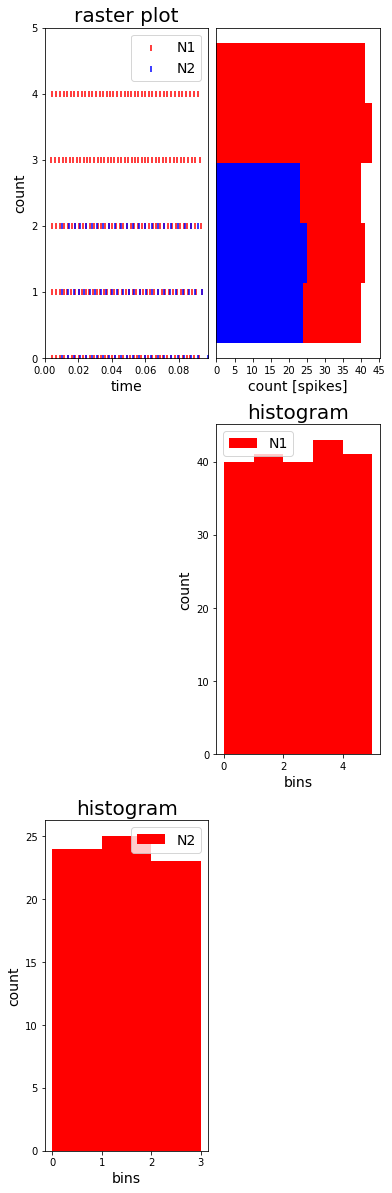
You can combine different plot in any kind of way. Just specifiy the location and size of the subplots you would want to have and then pass the respective subplots on to the different controller. When using more than one controller make sure to set show_immediately only to true in the controller that is called last.
… with matplotlib
# define plot structure BEFOREHAND
mainfig = plt.figure()
subfig1 = mainfig.add_subplot(321)
subfig2 = mainfig.add_subplot(322)
subfig3 = mainfig.add_subplot(324)
subfig4 = mainfig.add_subplot(325)
plt.subplots_adjust(left=0.125, right=0.9, bottom=0.1, top=4., wspace=0.05, hspace=0.2)
MyEventsModels = [spikemonN1, spikemonN2]
subgroup_labels = ['N1', 'N2']
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels,
mainfig=mainfig, subfig_rasterplot=subfig1, subfig_histogram = subfig2,
add_histogram=True, show_immediately=False)
DataModel_to_attr = [(spikemonN1, 'i')]
subgroup_labels = ['N1']
HC = Histogram(DataModel_to_attr=DataModel_to_attr, MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels, mainfig=mainfig, subfig=subfig3, show_immediately=False)
DataModel_to_attr = [(spikemonN2, 'i')]
subgroup_labels = ['N2']
HC = Histogram(DataModel_to_attr=DataModel_to_attr, MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels, mainfig=mainfig, subfig=subfig4, show_immediately=True)
… with pyqtgraph
# define plot structure BEFOREHAND
mainfig = pg.GraphicsWindow()
subfig1 = mainfig.addPlot(row=0, col=0)
subfig2 = mainfig.addPlot(row=0, col=1)
subfig2.setYLink(subfig1)
subfig3 = mainfig.addPlot(row=1, col=1)
subfig4 = mainfig.addPlot(row=2, col=0)
plt.subplots_adjust(left=0.125, right=0.9, bottom=0.1, top=4., wspace=0.05, hspace=0.2)
MyEventsModels = [spikemonN1, spikemonN2]
subgroup_labels = ['N1', 'N2']
RC = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels,
mainfig=mainfig, subfig_rasterplot=subfig1, subfig_histogram = subfig2, QtApp=QtApp,
backend='pyqtgraph', add_histogram=True, show_immediately=False)
DataModel_to_attr = [(spikemonN1, 'i')]
subgroup_labels = ['N1']
HC = Histogram(DataModel_to_attr=DataModel_to_attr, MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='pyqtgraph', mainfig=mainfig, subfig=subfig3, QtApp=QtApp,
show_immediately=False)
DataModel_to_attr = [(spikemonN1, 'i')]
subgroup_labels = ['N2']
HC = Histogram(DataModel_to_attr=DataModel_to_attr, MyPlotSettings=MyPlotSettings,
subgroup_labels=subgroup_labels,
backend='pyqtgraph', mainfig=mainfig, subfig=subfig4, QtApp=QtApp,
show_immediately=True)
Add second plot with a detailed view of a given plot¶
Create original plot of which you would like to have a detailed version as well TWICE
MyEventsModels = [spikemonN1, spikemonN2]
subgroup_labels = ['N1', 'N2']
mainfig = pg.GraphicsWindow()
subfig1 = mainfig.addPlot(row=0, col=0)
mainfig.nextRow()
subfig2 = mainfig.addPlot(row=1, col=0)
RC_org = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels,
mainfig=mainfig, subfig_rasterplot=subfig1,
QtApp=QtApp, backend='pyqtgraph', show_immediately=False)
RC_detail = Rasterplot(MyEventsModels=MyEventsModels, MyPlotSettings=MyPlotSettings, subgroup_labels=subgroup_labels,
mainfig=mainfig, subfig_rasterplot=subfig2,
QtApp=QtApp, backend='pyqtgraph', show_immediately=False)
RC_org.connect_detailed_subplot(filled_subplot_original_view=RC_org.viewer.subfig_rasterplot,
filled_subplot_detailed_view=RC_detail.viewer.subfig_rasterplot,
~